Note
Go to the end to download the full example code.
Knockoff aggregation on simulated data#
In this example, we show an example of variable selection using model-X Knockoffs introduced by Candès et al.[1]. A notable drawback of this procedure is the randomness associated with the knockoff generation process. This can result in unstable inference.
This example exhibits the two aggregation procedures described by Nguyen et al.[2] and Ren and Barber[3] to derandomize inference.
References#
Imports needed for this script#
import matplotlib.pyplot as plt
import numpy as np
from sklearn.utils import check_random_state
from hidimstat.data_simulation import simu_data
from hidimstat.knockoff_aggregation import knockoff_aggregation
from hidimstat.knockoffs import model_x_knockoff
from hidimstat.utils import cal_fdp_power
plt.rcParams.update({"font.size": 26})
# Number of observations
n_subjects = 500
# Number of variables
n_clusters = 500
# Correlation parameter
rho = 0.7
# Ratio of number of variables with non-zero coefficients over total
# coefficients
sparsity = 0.1
# Desired controlled False Discovery Rate (FDR) level
fdr = 0.1
seed = 45
n_bootstraps = 25
n_jobs = 10
runs = 20
rng = check_random_state(seed)
seed_list = rng.randint(1, np.iinfo(np.int32).max, runs)
def single_run(
n_subjects, n_clusters, rho, sparsity, fdr, n_bootstraps, n_jobs, seed=None
):
# Generate data
X, y, _, non_zero_index = simu_data(
n_subjects, n_clusters, rho=rho, sparsity=sparsity, seed=seed
)
# Use model-X Knockoffs [1]
mx_selection = model_x_knockoff(X, y, fdr=fdr, n_jobs=n_jobs, seed=seed)
fdp_mx, power_mx = cal_fdp_power(mx_selection, non_zero_index)
# Use p-values aggregation [2]
aggregated_ko_selection = knockoff_aggregation(
X,
y,
fdr=fdr,
n_bootstraps=n_bootstraps,
n_jobs=n_jobs,
gamma=0.3,
random_state=seed,
)
fdp_pval, power_pval = cal_fdp_power(aggregated_ko_selection, non_zero_index)
# Use e-values aggregation [1]
eval_selection = knockoff_aggregation(
X,
y,
fdr=fdr,
method="e-values",
n_bootstraps=n_bootstraps,
n_jobs=n_jobs,
gamma=0.3,
random_state=seed,
)
fdp_eval, power_eval = cal_fdp_power(eval_selection, non_zero_index)
return fdp_mx, fdp_pval, fdp_eval, power_mx, power_pval, power_eval
fdps_mx = []
fdps_pval = []
fdps_eval = []
powers_mx = []
powers_pval = []
powers_eval = []
for seed in seed_list:
fdp_mx, fdp_pval, fdp_eval, power_mx, power_pval, power_eval = single_run(
n_subjects, n_clusters, rho, sparsity, fdr, n_bootstraps, n_jobs, seed=seed
)
fdps_mx.append(fdp_mx)
fdps_pval.append(fdp_pval)
fdps_eval.append(fdp_eval)
powers_mx.append(fdp_mx)
powers_pval.append(power_pval)
powers_eval.append(power_eval)
# Plot FDP and Power distributions
fdps = [fdps_mx, fdps_pval, fdps_eval]
powers = [powers_mx, powers_pval, powers_eval]
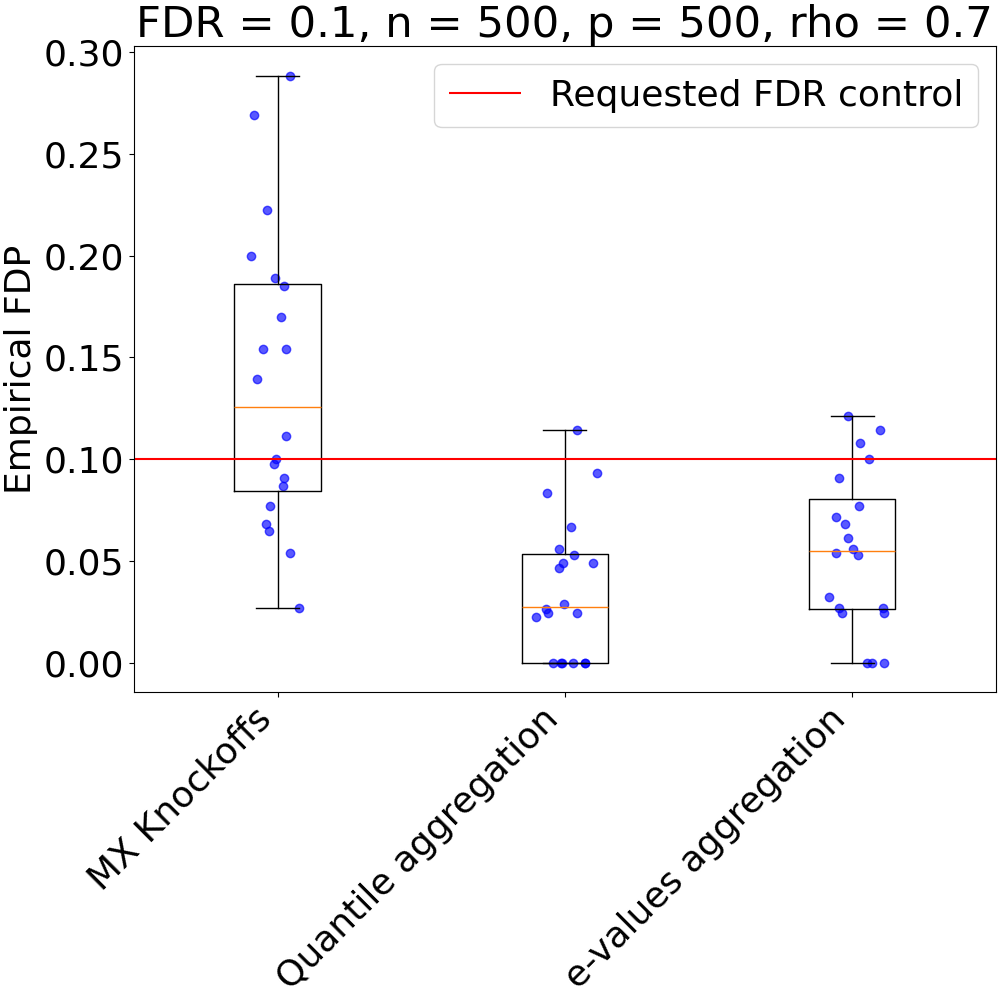
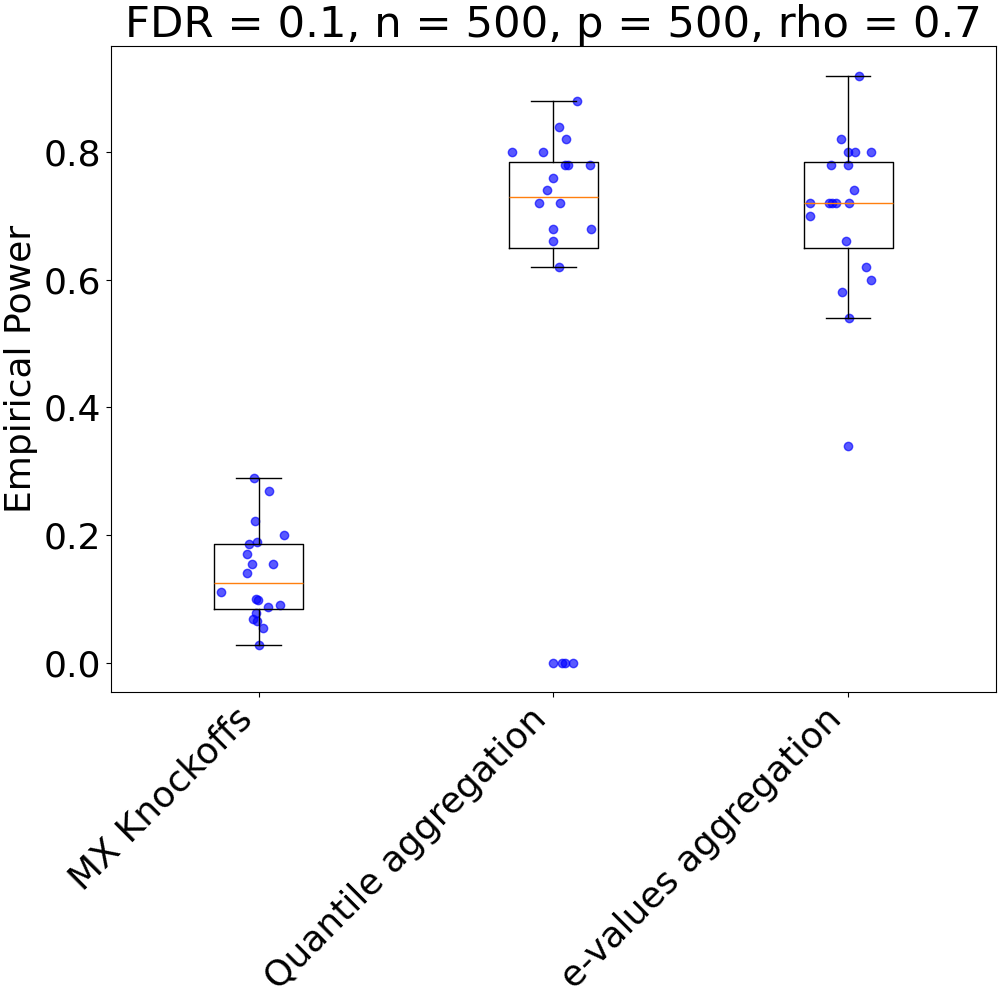
def plot_results(bounds, fdr, nsubjects, n_clusters, rho, power=False):
plt.figure(figsize=(10, 10), layout="constrained")
for nb in range(len(bounds)):
for i in range(len(bounds[nb])):
y = bounds[nb][i]
x = np.random.normal(nb + 1, 0.05)
plt.scatter(x, y, alpha=0.65, c="blue")
plt.boxplot(bounds, sym="")
if power:
plt.xticks(
[1, 2, 3],
["MX Knockoffs", "Quantile aggregation", "e-values aggregation"],
rotation=45,
ha="right",
)
plt.title(f"FDR = {fdr}, n = {nsubjects}, p = {n_clusters}, rho = {rho}")
plt.ylabel("Empirical Power")
else:
plt.hlines(fdr, xmin=0.5, xmax=3.5, label="Requested FDR control", color="red")
plt.xticks(
[1, 2, 3],
["MX Knockoffs", "Quantile aggregation", "e-values aggregation"],
rotation=45,
ha="right",
)
plt.title(f"FDR = {fdr}, n = {nsubjects}, p = {n_clusters}, rho = {rho}")
plt.ylabel("Empirical FDP")
plt.legend(loc="best")
plt.show()
plot_results(fdps, fdr, n_subjects, n_clusters, rho)
plot_results(powers, fdr, n_subjects, n_clusters, rho, power=True)
Total running time of the script: (5 minutes 13.519 seconds)
Estimated memory usage: 343 MB