8. Non-Cartesian 3D under-sampling#
In this notebook, you can play with the design parameters to regenerate different 3D sampling in-out patterns (so, we draw as many spiral arches as the number of shots). You can play with the number of shots by changing the under-sampling factor.
Author: Philippe Ciuciu (philippe.ciuciu@cea.fr) & Pierre-Antoine Comby (pierre-antoine.comby@ens-paris-saclay.fr)
Date: 01/30/2024 (dependence on MRI-NUFFT for 3D trajectories and NUFFT operators with density compensation)
Date: 04/02/2025 support GPUNUFFT with Pipe’s method for density compensation
Target: ATSI MSc students at Paris-Saclay University
import brainweb_dl as bwdl
import numpy as np
import matplotlib.pyplot as plt
import mrinufft
from mrinufft import display_2D_trajectory, display_3D_trajectory
from mrinufft import get_density, get_operator, check_backend
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-acq-recon-book/mri-acq-recon-book/venv/lib/python3.10/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
def _fit_grid(n_tiles):
"""Give the number of row and columns to optimally fit n_tiles."""
n_rows = int(np.sqrt(n_tiles))
n_cols = n_rows
while n_rows * n_cols < n_tiles:
if n_rows < n_cols:
n_rows += 1
else:
n_cols += 1
return n_rows, n_cols
def tile_view(
array,
axis=-1,
samples=-1,
n_rows=-1,
n_cols=-1,
img_w=3,
fig=None,
cmap="gray",
axis_label=None,
):
"""Plot a 3D array as a tiled grid of 2D images.
Parameters
----------
array: numpy.ndarray
A 3D array
axis: int
the axis on which to performs the sampling
samples: int or float, default: -1
If int, represent the number of equaly sample to take along axis.
If float, must be in (0, 1], and represent the sampling rate.
n_rows int, default=-1
n_cols int, default=-1
img_w: int
size in inches of a sample tile.
fig: Figure handle, default None
If None, a new figure is created.
cmap: matplotlib colormap.
color map to use.
Returns
-------
matplotlib.pyplot.figure
The figure handle
Raises
------
ValueError
If array is not 3D
ValueError
If both n_rows and n_cols are specified and cannot fit the samples.
"""
if array.ndim != 3:
raise ValueError("Only 3D array are supported.")
if axis < 0:
axis = 3 + axis
slicer = [slice(None), slice(None), slice(None)]
axis_label = axis_label or ["t", "x", "y", "z"][axis]
if samples == -1:
samples_loc = np.arange(array.shape[axis])
step = 1
elif isinstance(samples, int):
step = array.shape[axis] // (samples + 1)
samples_loc = np.arange(1, samples + 1) * step
elif isinstance(samples, float) and (0 < samples <= 1):
n_samples = int(array.shape[axis] * samples)
step = array.shape[axis] // n_samples
samples_loc = np.arange(0, n_samples) * step
else:
raise ValueError("Unsupported value for sample")
n_samples = len(samples_loc)
array_list = [array[(*slicer[:axis], s, *slicer[axis + 1 :])] for s in samples_loc]
if n_rows == -1 and n_cols != -1:
while n_rows * n_cols < n_samples:
n_rows += 1
elif n_rows != -1 and n_cols == -1:
while n_rows * n_cols < n_samples:
n_cols += 1
elif n_rows == -1 and n_cols == -1:
n_rows, n_cols = _fit_grid(n_samples)
elif n_rows != -1 and n_cols != -1 and n_rows * n_cols < n_samples:
raise ValueError("The grid is not big enough to fit all samples.")
aspect_ratio = array_list[0].shape[0] / array_list[0].shape[1]
fig = plt.figure(num=fig, figsize=(n_cols * img_w, n_rows * img_w * aspect_ratio))
gs = fig.add_gridspec(
n_rows,
n_cols,
hspace=0.01,
wspace=0.01,
)
axs_2d = gs.subplots(squeeze=False)
axs = axs_2d.flatten()
for i, img in enumerate(array_list):
ax = axs[i]
ax.axis("off")
if np.any(np.iscomplex(img)):
ax.imshow(abs(img), cmap=cmap, origin="lower")
else:
ax.imshow(img, cmap=cmap, origin="lower")
ax.text(
0.05,
0.95,
f"{axis_label}={step*i}",
ha="left",
va="top",
transform=ax.transAxes,
bbox=dict(boxstyle="square, pad=0", fc=fig.get_facecolor(), ec="none"),
)
# remove unused axis
for ax in axs[len(array_list) :]:
ax.remove()
return fig
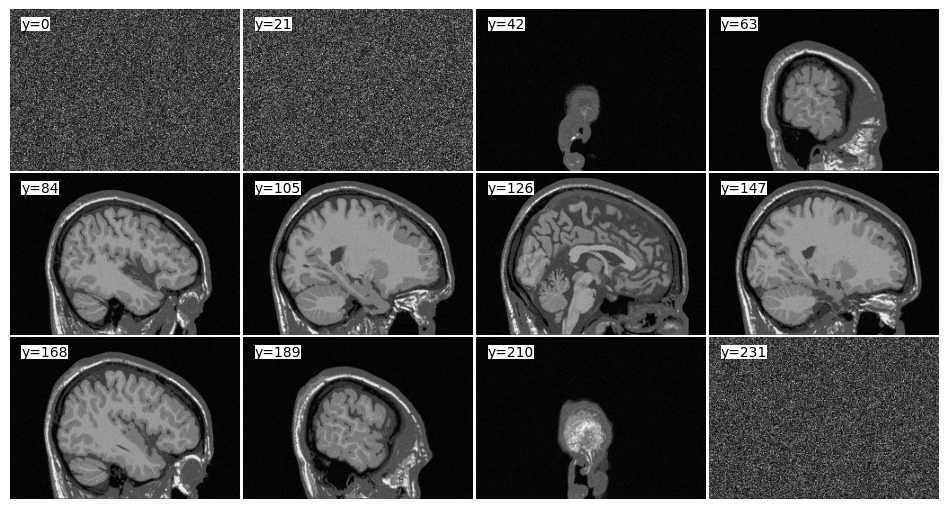
mri_3D = bwdl.get_mri(4, "T1").astype(np.float32)
print(mri_3D.shape)
# Axial views
#tile_view(mri_3D, samples=0.1, axis=0)
# Coronal views
#tile_view(mri_3D, samples=0.1, axis=1)
# Sagittal views
tile_view(mri_3D, samples=0.05, axis=-1)
(181, 256, 256)

def show_trajectory(trajectory, one_shot, figure_size):
ax = display_3D_trajectory(
trajectory, size=figure_size, one_shot=one_shot, per_plane=False
)
plt.tight_layout()
plt.subplots_adjust(bottom=0.1)
plt.show()
from mrinufft.trajectories import initialize_3D_cones
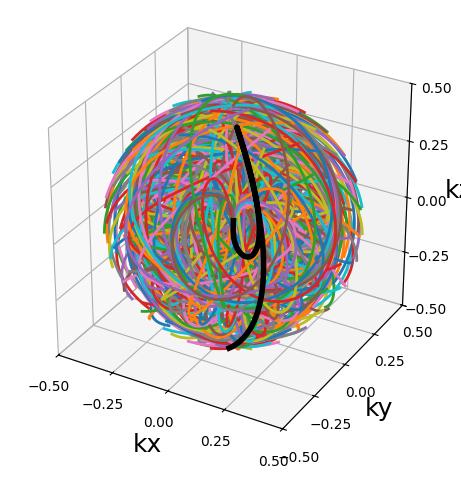
traj = initialize_3D_cones(256, 256).astype(np.float32)
show_trajectory(traj, 20, (15, 5))
# call to the ninufft operator without density compensation
#nufft = get_operator("finufft")(traj, mri_3D.shape, density=False)
# call to the gpunufft backend with density compensatioon based on Pipe's method
nufft = get_operator("gpunufft")(traj, mri_3D.shape, density=True)
kspace = nufft.op(mri_3D)
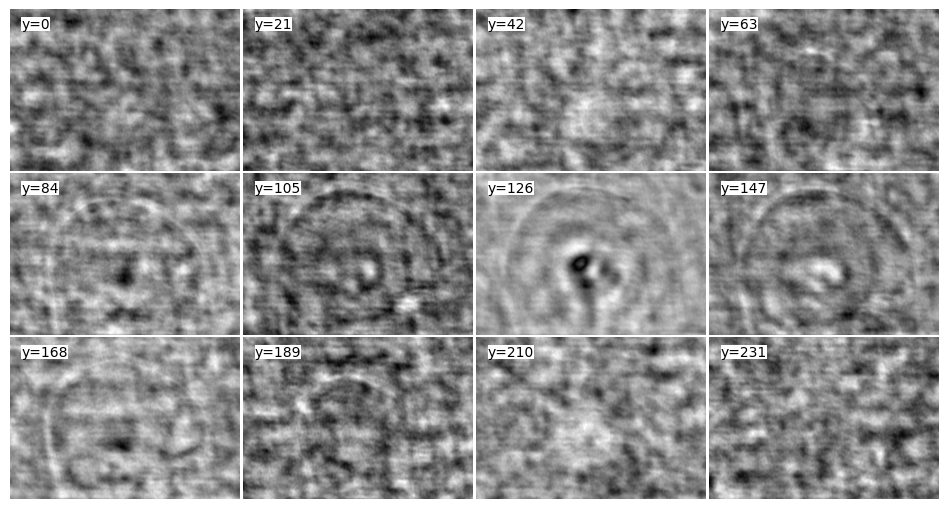
adjoint_3Dcones = nufft.adj_op(kspace)
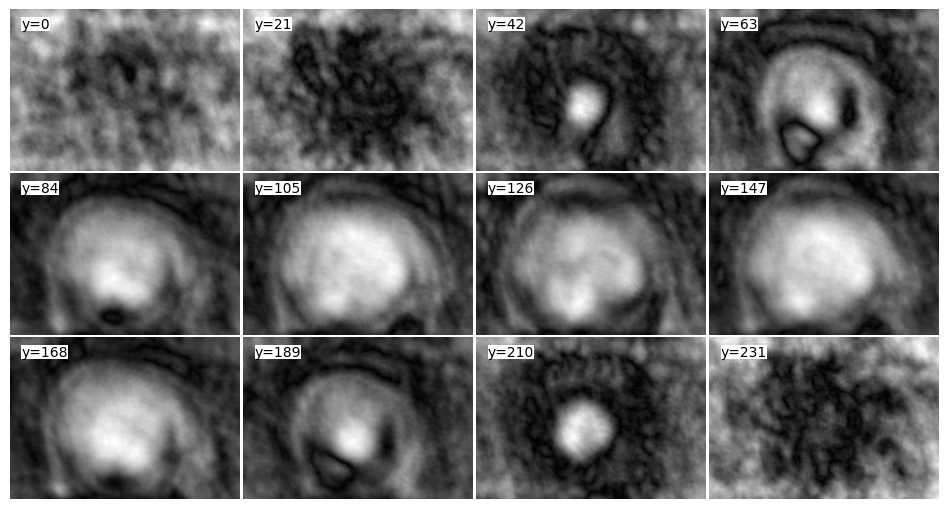
tile_view(adjoint_3Dcones, samples=0.05, axis=-1)
#cell_weights = get_density("cell_count", traj, shape=mri_3D.shape, osf=2.0)
#nufft_cell_counting = get_operator("finufft")(
# traj, shape=mri_3D.shape, density=cell_weights, upsampfac=2.0
#)
#dc_adjoint_3Dcones = nufft_cell_counting.adj_op(kspace)
#tile_view(dc_adjoint_3Dcones, samples=0.05, axis=-1)
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Cell In[5], line 10
5 show_trajectory(traj, 20, (15, 5))
7 # call to the ninufft operator without density compensation
8 #nufft = get_operator("finufft")(traj, mri_3D.shape, density=False)
9 # call to the gpunufft backend with density compensatioon based on Pipe's method
---> 10 nufft = get_operator("gpunufft")(traj, mri_3D.shape, density=True)
12 kspace = nufft.op(mri_3D)
13 adjoint_3Dcones = nufft.adj_op(kspace)
File ~/gpu_mind_runner/_work/mri-acq-recon-book/mri-acq-recon-book/venv/lib/python3.10/site-packages/mrinufft/operators/interfaces/gpunufft.py:422, in MRIGpuNUFFT.__init__(self, samples, shape, n_coils, n_batchs, n_trans, density, smaps, squeeze_dims, eps, **kwargs)
420 self.n_batchs = n_batchs
421 self.squeeze_dims = squeeze_dims
--> 422 self.compute_density(density)
423 self.compute_smaps(smaps)
424 self.raw_op = RawGpuNUFFT(
425 samples=self.samples,
426 shape=self.shape,
(...)
431 **kwargs,
432 )
File ~/gpu_mind_runner/_work/mri-acq-recon-book/mri-acq-recon-book/venv/lib/python3.10/site-packages/mrinufft/operators/base.py:332, in FourierOperatorBase.compute_density(self, method)
326 if self._density_method is None:
327 self._density_method = lambda samples, shape: method(
328 samples,
329 shape,
330 **kwargs,
331 )
--> 332 self._density = method(self.samples, self.shape, **kwargs)
File ~/gpu_mind_runner/_work/mri-acq-recon-book/mri-acq-recon-book/venv/lib/python3.10/site-packages/mrinufft/density/utils.py:35, in flat_traj.<locals>.decorator.<locals>.wrapper(*args, **kwargs)
33 args = list(args)
34 args[0] = proper_trajectory(args[0], normalize=normalize)
---> 35 return func(*args, **kwargs)
File ~/gpu_mind_runner/_work/mri-acq-recon-book/mri-acq-recon-book/venv/lib/python3.10/site-packages/mrinufft/density/nufft_based.py:27, in pipe(traj, shape, backend, **kwargs)
25 nufft_class = get_operator(backend)
26 if hasattr(nufft_class, "pipe"):
---> 27 return nufft_class.pipe(traj, shape, **kwargs)
28 raise ValueError("backend does not have pipe iterations method.")
File ~/gpu_mind_runner/_work/mri-acq-recon-book/mri-acq-recon-book/venv/lib/python3.10/site-packages/mrinufft/operators/interfaces/gpunufft.py:612, in MRIGpuNUFFT.pipe(cls, kspace_loc, volume_shape, num_iterations, osf, normalize, **kwargs)
610 test_op = MRIGpuNUFFT(samples=kspace_loc, shape=original_shape, **kwargs)
611 test_im = np.ones(original_shape, dtype=np.complex64)
--> 612 test_im_recon = test_op.adj_op(density_comp * test_op.op(test_im))
613 density_comp /= np.mean(np.abs(test_im_recon))
614 return density_comp.squeeze()
File ~/gpu_mind_runner/_work/mri-acq-recon-book/mri-acq-recon-book/venv/lib/python3.10/site-packages/mrinufft/_array_compat.py:103, in with_numpy_cupy.<locals>.wrapper(*args, **kwargs)
100 xp = get_array_module(leading_arg)
102 # convert all to cupy / numpy according to data arg device
--> 103 args, kwargs = _to_numpy_cupy(args, kwargs, leading_arg)
105 # run function
106 ret_ = fun(*args, **kwargs)
File ~/gpu_mind_runner/_work/mri-acq-recon-book/mri-acq-recon-book/venv/lib/python3.10/site-packages/mrinufft/_array_compat.py:251, in _to_numpy_cupy(args, kwargs, leading_argument)
249 if is_cuda_array(leading_argument) and CUPY_AVAILABLE:
250 return _to_cupy(*args, **kwargs)
--> 251 elif is_cuda_tensor(leading_argument) and CUPY_AVAILABLE:
252 return _to_cupy(*args, **kwargs)
253 else:
File ~/gpu_mind_runner/_work/mri-acq-recon-book/mri-acq-recon-book/venv/lib/python3.10/site-packages/mrinufft/operators/interfaces/utils/gpu_utils.py:39, in is_cuda_tensor(var)
37 def is_cuda_tensor(var):
38 """Check if var is a CUDA tensor."""
---> 39 return isinstance(var, torch.Tensor) and var.is_cuda
NameError: name 'torch' is not defined
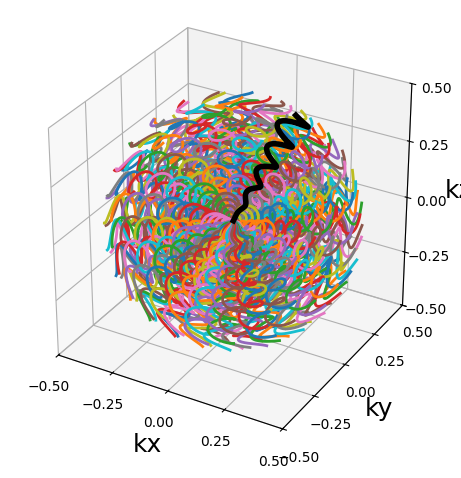
from mrinufft.trajectories import initialize_3D_floret
traj = initialize_3D_floret(256, 256).astype(np.float32)
show_trajectory(traj, 10, (15, 5))
# call to the ninufft operator without density compensation
#nufft = get_operator("finufft")(traj, mri_3D.shape, density=False)
# call to the gpunufft backend with density compensatioon based on Pipe's method
nufft = get_operator("gpunufft")(traj, mri_3D.shape, density=True)
kspace = nufft.op(mri_3D)
adjoint_3Dfloret = nufft.adj_op(kspace)
tile_view(adjoint_3Dfloret, samples=0.05, axis=-1)
#cell_weights = get_density("cell_count", traj, shape=mri_3D.shape, osf=2.0)
#nufft_cell_counting = get_operator("finufft")(
# traj, shape=mri_3D.shape, density=cell_weights, upsampfac=2.0
#)
#dc_adjoint_3Dfloret = nufft_cell_counting.adj_op(kspace)
#tile_view(dc_adjoint_3Dfloret, samples=0.05, axis=-1)
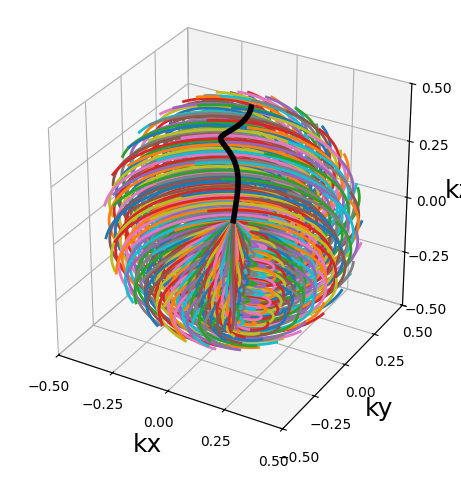

from mrinufft.trajectories import initialize_3D_wave_caipi
traj = initialize_3D_wave_caipi(256, 256).astype(np.float32)
show_trajectory(traj, 10, (15, 5))
# call to the ninufft operator without density compensation
#nufft = get_operator("finufft")(traj, mri_3D.shape, density=False)
# call to the gpunufft backend with density compensatioon based on Pipe's method
nufft = get_operator("gpunufft")(traj, mri_3D.shape, density=True)
kspace = nufft.op(mri_3D)
adjoint_3Dwave_caipi = nufft.adj_op(kspace)
tile_view(adjoint_3Dwave_caipi, samples=0.05, axis=-1)
#voronoi_weights = get_density("voronoi", traj)
#nufft3D_voronoi = get_operator("finufft")(
# traj, shape=mri_3D.shape, density=voronoi_weights
#)
#dc_adjoint_3Dwave_caipi = nufft3D_voronoi.adj_op(kspace)
#cell_weights = get_density("cell_count", traj, shape=mri_3D.shape, osf=2.0)
##nufft_cell_counting = get_operator("finufft")(
# traj, shape=mri_3D.shape, density=cell_weights, upsampfac=2.0
#)
#dc_adjoint_3Dwave_caipi = nufft_cell_counting.adj_op(kspace)
#tile_view(dc_adjoint_3Dwave_caipi, samples=0.05, axis=-1)
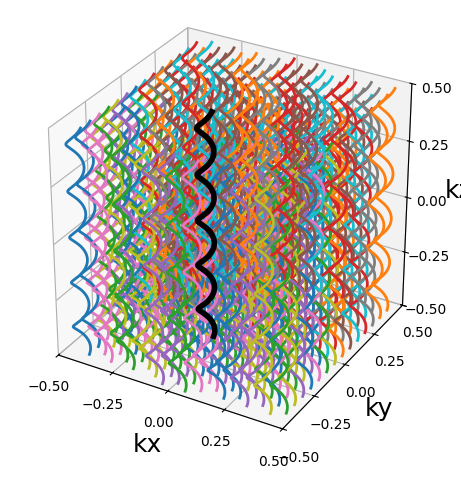

from mrinufft.trajectories import initialize_3D_seiffert_spiral
traj = initialize_3D_seiffert_spiral(256, 256).astype(np.float32)
show_trajectory(traj, 10, (15, 5))
# call to the ninufft operator without density compensation
#nufft = get_operator("finufft")(traj, mri_3D.shape, density=False)
# call to the gpunufft backend with density compensatioon based on Pipe's method
nufft = get_operator("gpunufft")(traj, mri_3D.shape, density=True)
kspace = nufft.op(mri_3D)
adjoint_3D_seiffert_spiral = nufft.adj_op(kspace)
tile_view(adjoint_3D_seiffert_spiral, samples=0.05, axis=-1)