Note
Go to the end to download the full example code or to run this example in your browser via Binder.
Sensitivity maps estimation#
This example demonstrates how to estimate coil sensitivity maps from non-Cartesian k-space data using different methods provided in the :mrinufft:`mrinufft.extras.smaps` module. We will simulate k-space data from a known MRI image and coil sensitivity maps, and then estimate the sensitivity maps using the ESPIRiT method [espirit] and a low-frequency calibration method [sense]. We will visualize the estimated sensitivity maps and compare them to the actual sensitivity maps used in the simulation.
Imports#
import matplotlib.pyplot as plt
import numpy as np
from brainweb_dl import get_mri
from sigpy.mri.sim import birdcage_maps
from mrinufft import get_operator
from mrinufft.extras.smaps import get_smaps
from mrinufft.trajectories import initialize_3D_floret
import cupy as cp
import os
Function to display imgs
def show_maps(imgs):
"""Display 4D sensitivity maps in a 4x3 figure layout."""
n_coils, nx, ny, nz = imgs.shape
fig, axes = plt.subplots(4, 3, figsize=(15, 20))
imgs = np.abs(imgs)
for i in range(n_coils):
axes[i, 0].imshow(imgs[i, nx // 2, :, :], vmax=imgs.max(), vmin=imgs.min())
axes[i, 1].imshow(imgs[i, :, ny // 2, :], vmax=imgs.max(), vmin=imgs.min())
axes[i, 2].imshow(imgs[i, :, :, nz // 2], vmax=imgs.max(), vmin=imgs.min())
axes[i, 0].set_title(f"Coil {i+1} - YZ plane")
axes[i, 1].set_title(f"Coil {i+1} - XZ plane")
axes[i, 2].set_title(f"Coil {i+1} - XY plane")
plt.show()
return fig
BACKEND = os.environ.get("MRINUFFT_BACKEND", "cufinufft")
Get MRI data, 3D FLORET trajectory, and simulate k-space data
samples_loc = initialize_3D_floret(Nc=16 * 16, Ns=256)
mri = get_mri(0)[::2, ::2, ::2][::-1, ::-1] # Load and downsample MRI data for speed
n_coils = 4
actual_smaps = birdcage_maps(
(n_coils, *mri.shape), dtype=np.complex64
) # Generate birdcage sensitivity maps
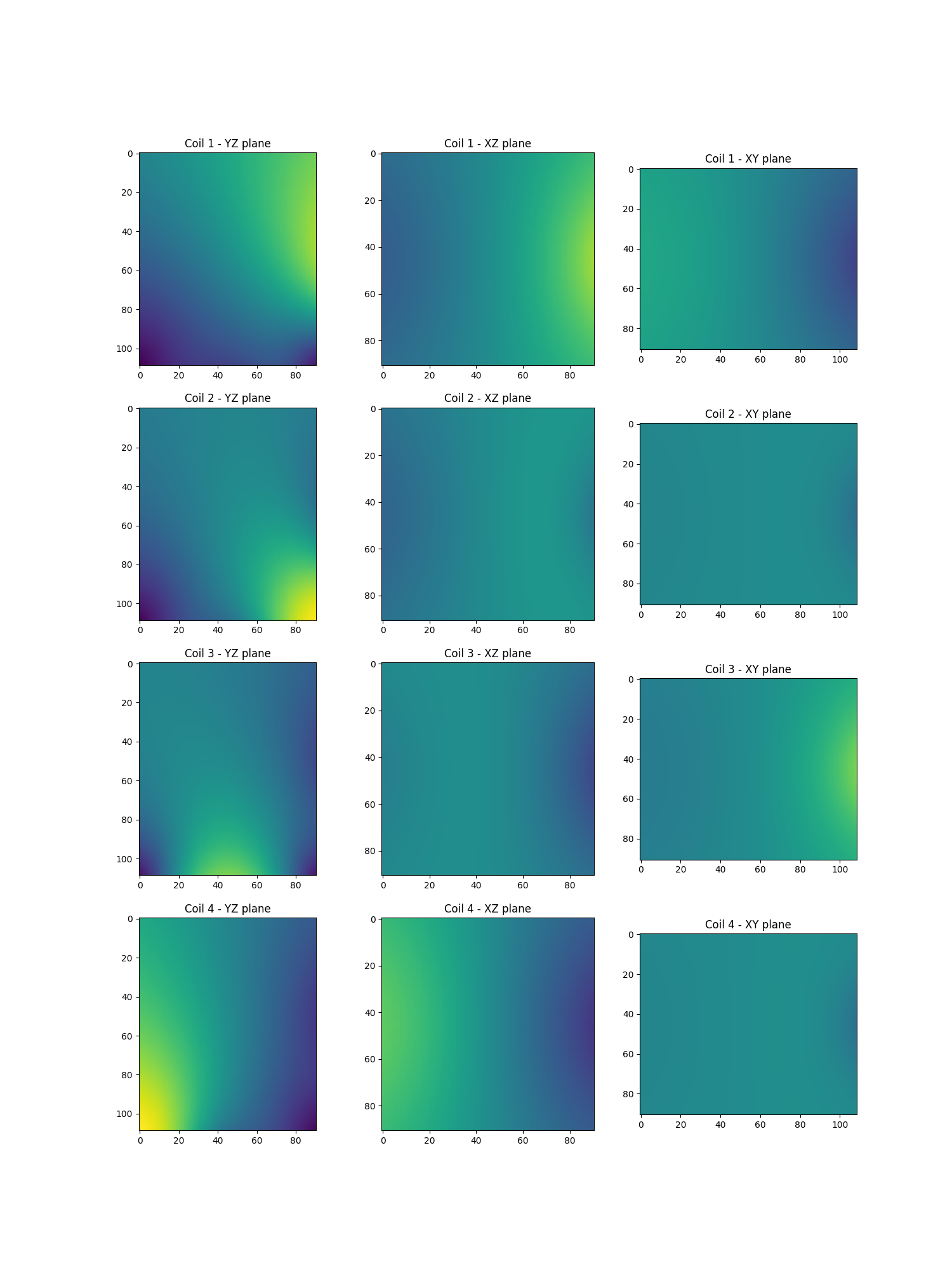
Show the sensitivity maps
show_maps(actual_smaps)
<Figure size 1500x2000 with 12 Axes>
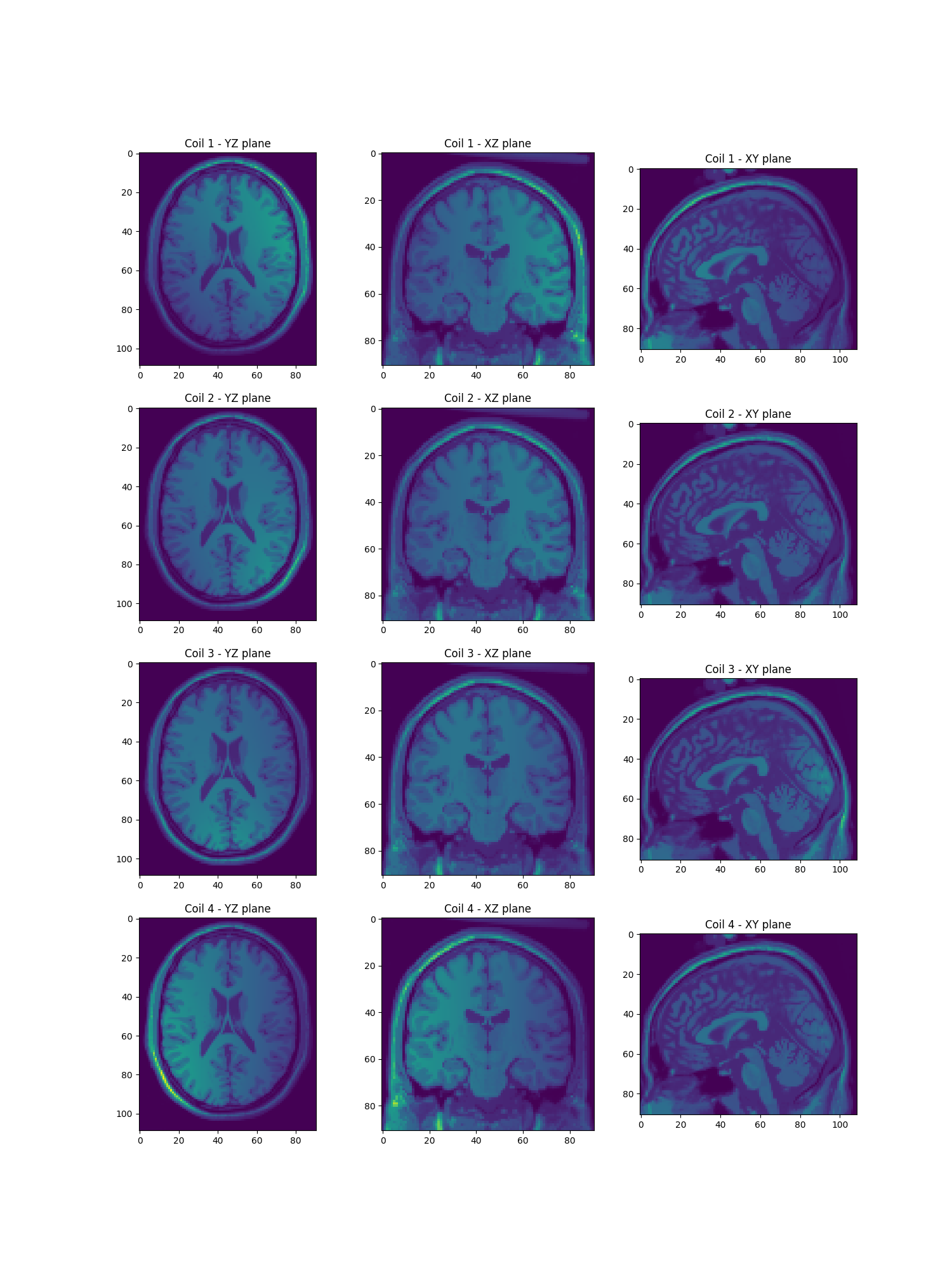
Show the per channel images
per_ch_mri = mri[None, ...] * actual_smaps # Generate per-coil MRI data
show_maps(per_ch_mri)
<Figure size 1500x2000 with 12 Axes>
Simulate k-space data
forward_op = get_operator(BACKEND)(
samples_loc, shape=mri.shape, n_coils=n_coils, density=True
)
kspace_data = forward_op.op(per_ch_mri) # Simulate k-space data
if str.lower(BACKEND) in ["gpunufft", "cufinufft"]:
# GPU exists, run on GPU
import cupy as cp
kspace_data = cp.asarray(kspace_data, dtype=cp.complex64)
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:67: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:72: UserWarning: Samples will be rescaled to [-0.5, 0.5), assuming they were in [-pi, pi)
warnings.warn(
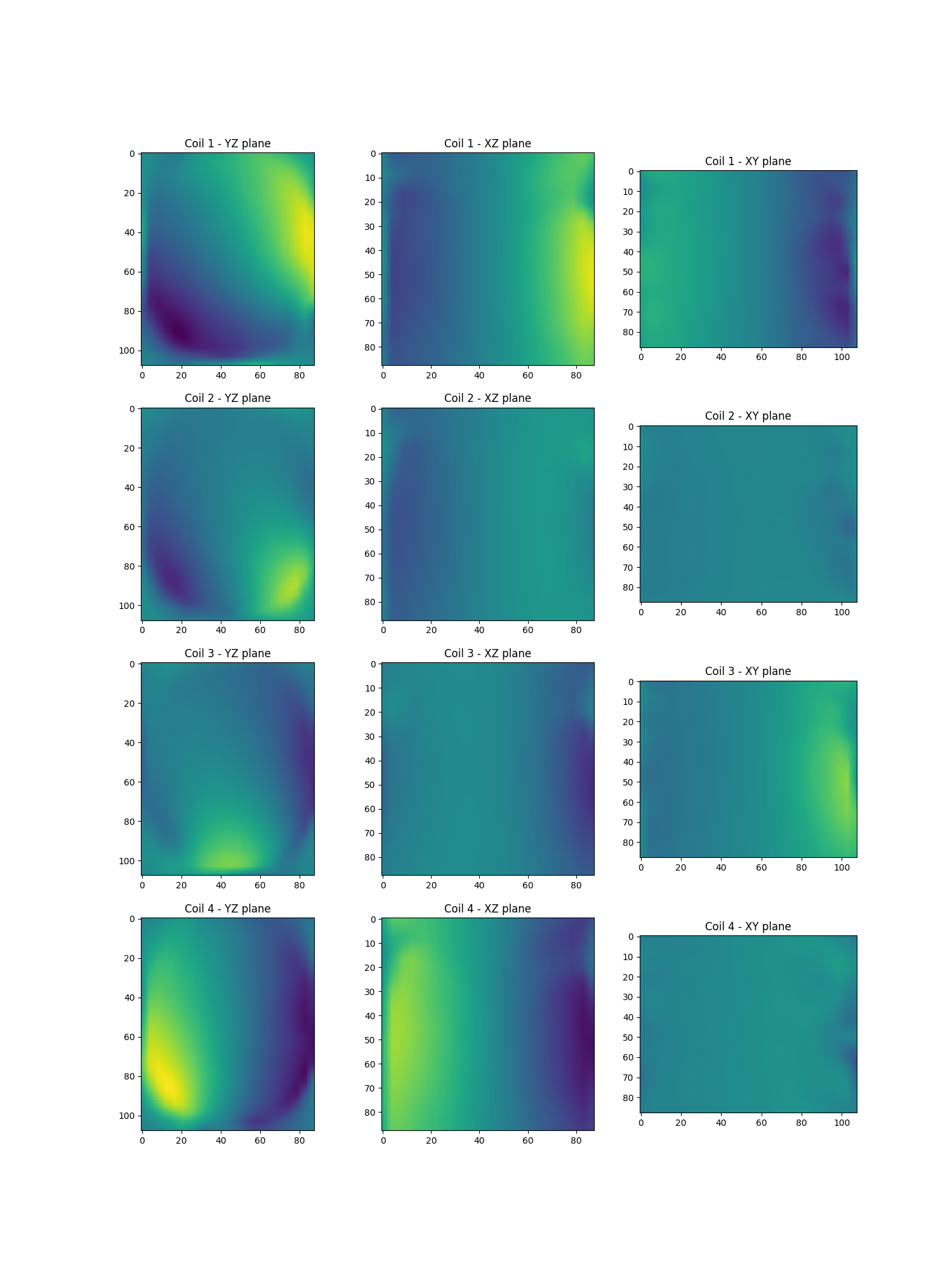
Estimate sensitivity maps using ESPIRiT
Smaps = get_smaps("espirit")(
samples_loc,
mri.shape,
kspace_data=kspace_data,
density=forward_op.density,
backend=BACKEND,
decim=4,
)
show_maps(Smaps.get())
0%| | 0/10 [00:00<?, ?it/s]
10%|█ | 1/10 [00:00<00:01, 5.44it/s]
20%|██ | 2/10 [00:00<00:01, 6.91it/s]
30%|███ | 3/10 [00:00<00:00, 7.50it/s]
40%|████ | 4/10 [00:00<00:00, 7.61it/s]
50%|█████ | 5/10 [00:00<00:00, 7.84it/s]
60%|██████ | 6/10 [00:00<00:00, 7.99it/s]
70%|███████ | 7/10 [00:00<00:00, 7.97it/s]
80%|████████ | 8/10 [00:01<00:00, 7.95it/s]
90%|█████████ | 9/10 [00:01<00:00, 8.12it/s]
100%|██████████| 10/10 [00:01<00:00, 8.19it/s]
100%|██████████| 10/10 [00:01<00:00, 7.81it/s]
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/operators/base.py:1061: UserWarning: Lipschitz constant did not converge
warnings.warn("Lipschitz constant did not converge")
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_array_compat.py:248: UserWarning: data is on gpu, it will be moved to CPU.
warnings.warn("data is on gpu, it will be moved to CPU.")
<Figure size 1500x2000 with 12 Axes>
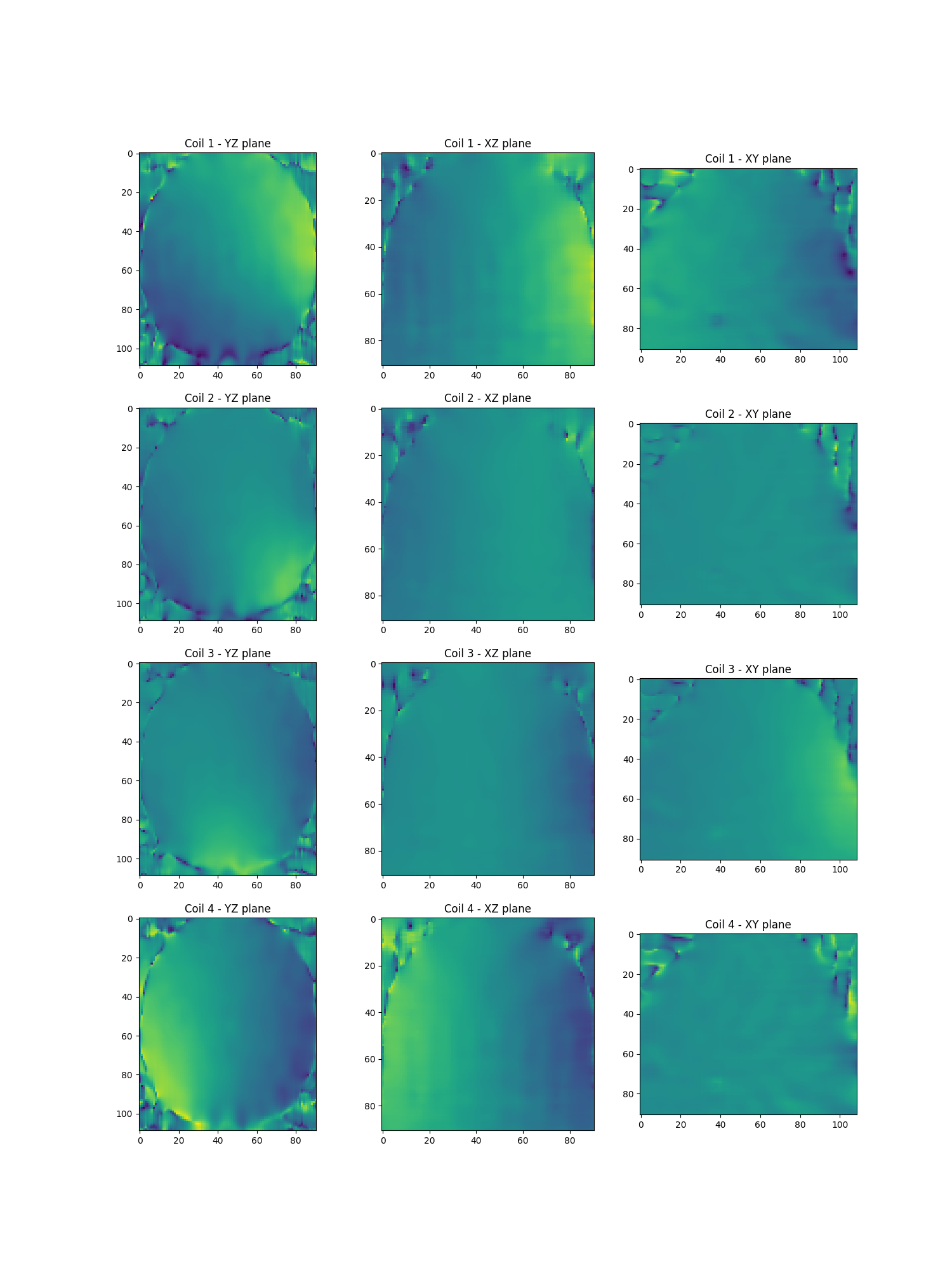
Estimate the sensitivity map using low-frequency calibration
Smaps = get_smaps("low_frequency")(
samples_loc,
mri.shape,
kspace_data=kspace_data,
density=forward_op.density,
backend=BACKEND,
)
show_maps(Smaps.get())
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:67: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
0%| | 0/10 [00:00<?, ?it/s]
10%|█ | 1/10 [00:00<00:01, 5.33it/s]
20%|██ | 2/10 [00:00<00:01, 5.81it/s]
30%|███ | 3/10 [00:00<00:01, 5.94it/s]
40%|████ | 4/10 [00:00<00:01, 5.72it/s]
50%|█████ | 5/10 [00:00<00:00, 5.60it/s]
60%|██████ | 6/10 [00:01<00:00, 5.53it/s]
70%|███████ | 7/10 [00:01<00:00, 5.49it/s]
80%|████████ | 8/10 [00:01<00:00, 5.46it/s]
90%|█████████ | 9/10 [00:01<00:00, 5.44it/s]
100%|██████████| 10/10 [00:01<00:00, 5.43it/s]
100%|██████████| 10/10 [00:01<00:00, 5.53it/s]
<Figure size 1500x2000 with 12 Axes>
References#
Loubna El Gueddari, C. Lazarus, H Carrié, A. Vignaud, Philippe Ciuciu. Self-calibrating nonlinear reconstruction algorithms for variable density sampling and parallel reception MRI. 10th IEEE Sensor Array and Multichannel Signal Processing workshop, Jul 2018, Sheffield, United Kingdom. ⟨hal-01782428v1⟩
Uecker M, Lai P, Murphy MJ, Virtue P, Elad M, Pauly JM, Vasanawala SS, Lustig M. ESPIRiT–an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA. Magn Reson Med. 2014 Mar;71(3):990-1001. doi: 10.1002/mrm.24751. PMID: 23649942; PMCID: PMC4142121.
Total running time of the script: (0 minutes 10.358 seconds)
