initialize_3D_floret#
- mrinufft.trajectories.trajectory3D.initialize_3D_floret(Nc: int, Ns: int, in_out: bool = False, nb_revolutions: float = 1, spiral: str | float = 'fermat', cone_tilt: str | float = 'golden', max_angle: float = 1.5707963267948966, axes: tuple[int, ...] = (2,)) ndarray[tuple[int, ...], dtype[_ScalarType_co]][source]#
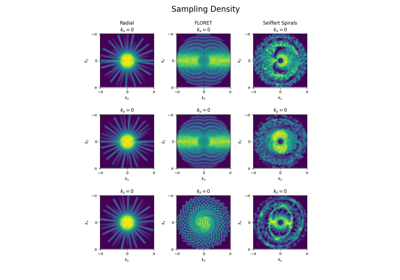
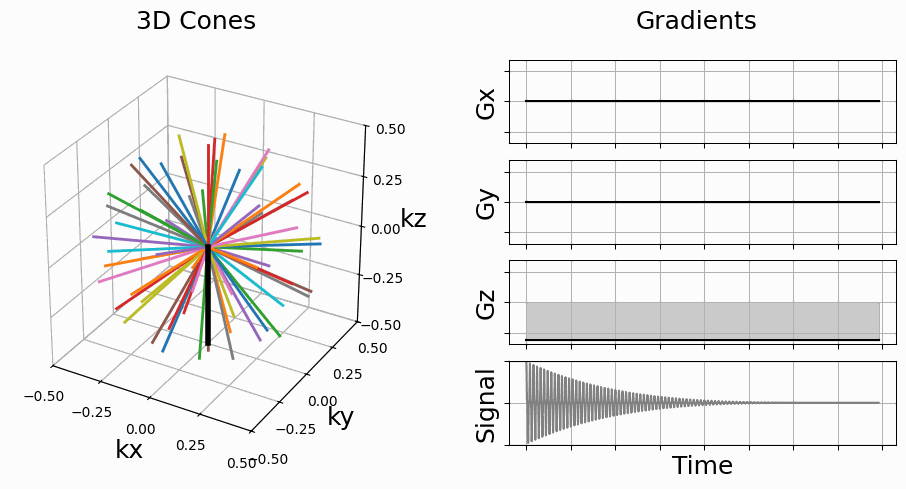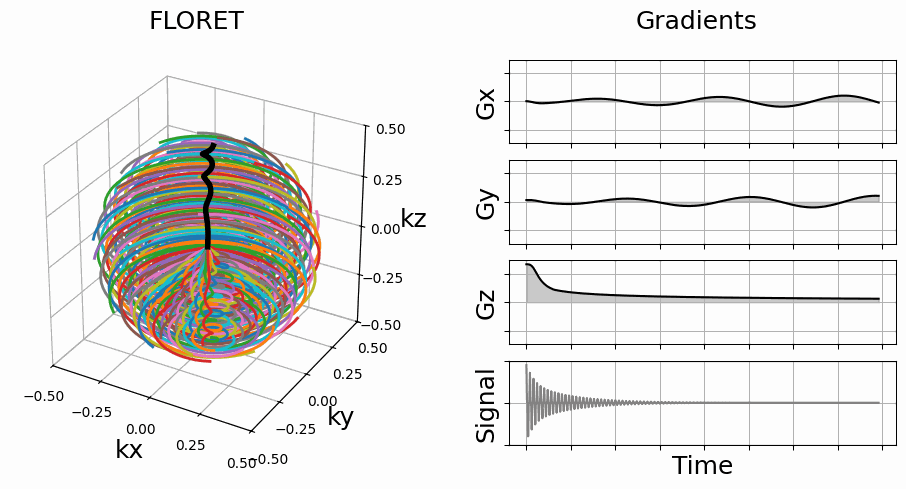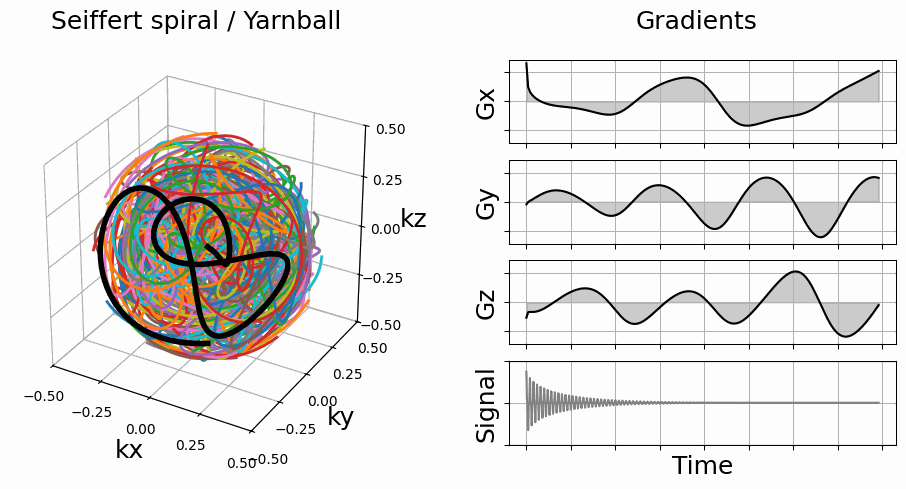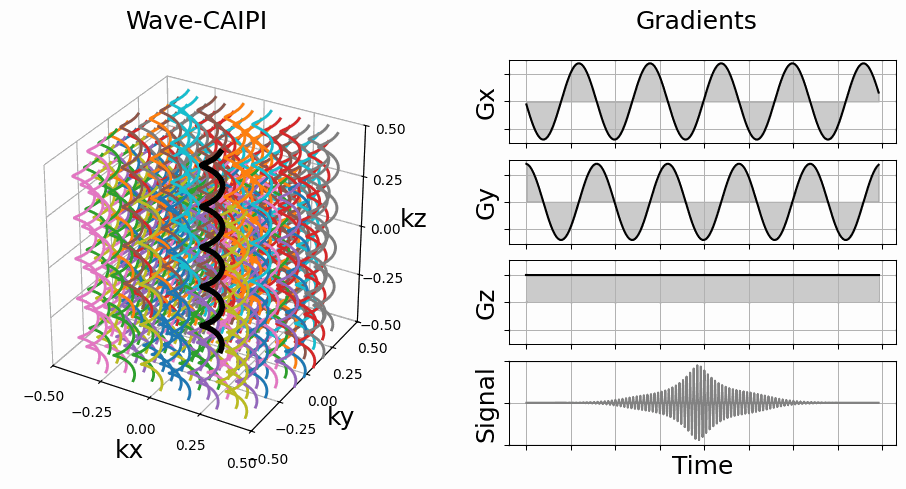
Initialize 3D trajectories with FLORET.
This implementation is based on the work from [Pip+11]. The acronym FLORET stands for Fermat Looped, Orthogonally Encoded Trajectories. It consists of Fermat spirals folded into 3D cones along one or several axes.
- Parameters:
Nc (int) – Number of shots
Ns (int) – Number of samples per shot
in_out (bool, optional) – Whether to start from the center or not, by default False
nb_revolutions (float, optional) – Number of revolutions of the spirals, by default 1
spiral (str, float, optional) – Spiral type, by default “fermat”
cone_tilt (str, float, optional) – Tilt of the cones around the \(k_z\)-axis, by default “golden”
max_angle (float, optional) – Maximum polar angle starting from the \(k_x-k_y\) plane, by default pi / 2
axes (tuple, optional) – Axes over which cones are created, by default (2,)
- Returns:
3D FLORET trajectory
- Return type:
NDArray
References
[Pip+11]Pipe, James G., Nicholas R. Zwart, Eric A. Aboussouan, Ryan K. Robison, Ajit Devaraj, and Kenneth O. Johnson. “A new design and rationale for 3D orthogonally oversampled k‐space trajectories.” Magnetic resonance in medicine 66, no. 5 (2011): 1303-1311.