Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Subspace NUFFT Operator#
An example to show how to setup a subspace NUFFT operator.
This example shows how to use the Subspace NUFFT operator to acquire data and reconstruct an image (or a volume for 3D imaging) while projecting onto a low rank spatiotemporal subspace. This can be useful e.g., for dynamic multi-contrast MRI. Hereafter a 2D spiral trajectory is used for demonstration.
import matplotlib.pyplot as plt
import numpy as np
from mrinufft import display_2D_trajectory
Data preparation#
Image loading#
For realistic a 2D image we will use the BrainWeb dataset,
installable using pip install brainweb-dl.
from mrinufft.extras import get_brainweb_map
M0, T1, T2 = get_brainweb_map(0)
M0 = np.flip(M0, axis=(0, 1, 2))[90]
T1 = np.flip(T1, axis=(0, 1, 2))[90]
T2 = np.flip(T2, axis=(0, 1, 2))[90]
fig1, ax1 = plt.subplots(1, 3)
im0 = ax1[0].imshow(M0, cmap="gray")
ax1[0].axis("off"), ax1[0].set_title("M0 [a.u.]")
fig1.colorbar(im0, ax=ax1[0], fraction=0.046, pad=0.04)
im1 = ax1[1].imshow(T1, cmap="magma")
ax1[1].axis("off"), ax1[1].set_title("T1 [ms]")
fig1.colorbar(im1, ax=ax1[1], fraction=0.046, pad=0.04)
im2 = ax1[2].imshow(T2, cmap="viridis")
ax1[2].axis("off"), ax1[2].set_title("T2 [ms]")
fig1.colorbar(im2, ax=ax1[2], fraction=0.046, pad=0.04)
plt.tight_layout()
plt.show()
![M0 [a.u.], T1 [ms], T2 [ms]](../../_images/sphx_glr_example_subspace_001.png)
Sequence Parameters#
As an example, we simulate a simple monoexponential Spin Echo acquisition. We assume that k-space center is sampled at each TE (as in spiral or radial imaging) and a constant flip angle train is constant set to 180 degrees. In this way, we obtain an image for each of the shots in the echo train.
Subspace Generation#
Here, we generate temporal subspace basis by performing the Singular Value Decomposition of an ensemble of simulated signals corresponding to the same T1-T2 range of our object.
def make_grid(T1range, T2range, natoms=100):
"""Prepare parameter grid for basis estimation."""
# Create linear grid
T1grid = np.linspace(T1range[0], T1range[1], num=natoms, dtype=np.float32)
T2grid = np.linspace(T2range[0], T2range[1], num=natoms, dtype=np.float32)
# Cartesian product
T1grid, T2grid = np.meshgrid(T1grid, T2grid)
return T1grid.ravel(), T2grid.ravel()
def estimate_subspace_basis(train_data, ncoeff=4):
"""Estimate subspace data via SVD of simulated training data."""
# Perform svd
_, _, basis = np.linalg.svd(train_data, full_matrices=False)
# Calculate basis (retain only ncoeff coefficients)
basis = basis[:ncoeff]
return basis
# Get range from tissues
T1range = (T1.min() + 1, T1.max()) # [ms]
T2range = (T2.min() + 1, T2.max()) # [ms]
# Prepare tissue grid
T1grid, T2grid = make_grid(T1range, T2range)
# Calculate training data
train_data = fse_simulation(1.0, T1grid, T2grid, TE, TR).astype(np.float32)
# Calculate basis
basis = estimate_subspace_basis(train_data.T)
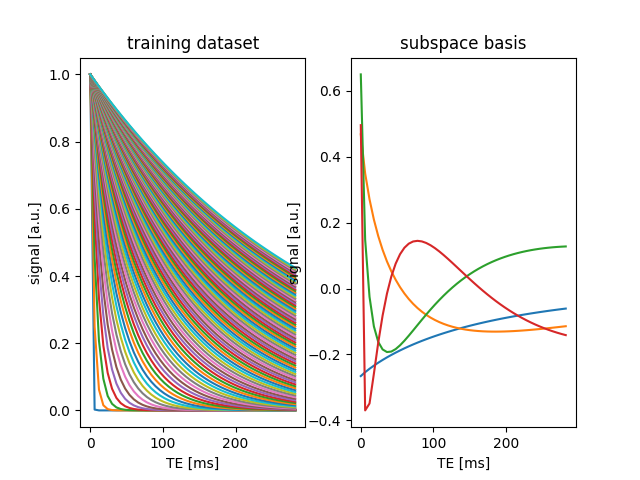
fig2, ax2 = plt.subplots(1, 2)
ax2[0].plot(TE, train_data[:, ::100]), ax2[0].set(
xlabel="TE [ms]", ylabel="signal [a.u.]"
), ax2[0].set_title("training dataset")
ax2[1].plot(TE, basis.T), ax2[1].set(xlabel="TE [ms]", ylabel="signal [a.u.]"), ax2[
1
].set_title("subspace basis")
plt.show()
Here, we simulate Brainweb FSE data with the same sequence parameters as those used for the subspace estimation.
mri_data = fse_simulation(M0, T1, T2, TE, TR).astype(np.float32)
mri_data = np.ascontiguousarray(mri_data)
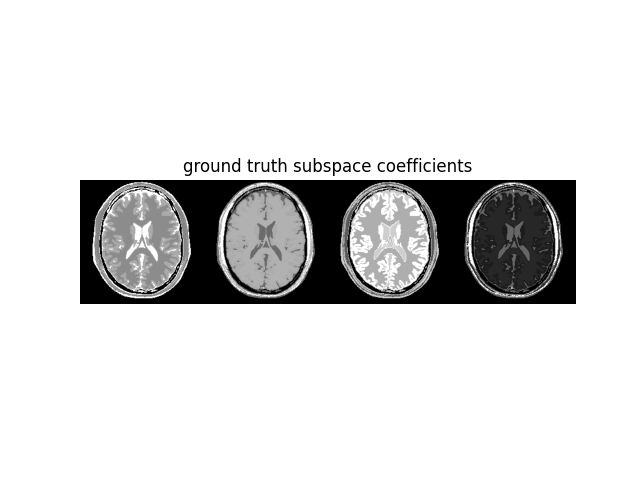
# Ground truth subspace coefficients
ground_truth = (mri_data.T @ basis.T).T
ground_truth = np.ascontiguousarray(ground_truth)
ground_truth_display = np.concatenate(
[abs(coeff) / abs(coeff).max() for coeff in ground_truth], axis=-1
)
plt.figure()
plt.imshow(ground_truth_display, cmap="gray"), plt.axis("off"), plt.title(
"ground truth subspace coefficients"
)
plt.show()
Trajectory generation#
from mrinufft import initialize_2D_spiral
from mrinufft.density import voronoi
samples = initialize_2D_spiral(
Nc=ETL * 16, Ns=1200, nb_revolutions=10, tilt="mri-golden"
)
# assume trajectory is reordered as (ncontrasts, nshots_per_contrast, nsamples_per_shot, ndims)
# with contrast axis sorted by ascending TEs
samples = samples.reshape(ETL, 16, *samples.shape[1:])
# compute density compensation
density = voronoi(samples)
density = density.reshape(ETL, 16, samples.shape[-2])
display_2D_trajectory(samples.reshape(-1, *samples.shape[2:]))
<Axes: xlabel='kx', ylabel='ky'>
Operator setup#
from mrinufft import get_operator
from mrinufft.operators import MRISubspace
# Generate standard NUFFT operator
nufft = get_operator("finufft")(
samples=samples,
shape=mri_data.shape[-2:],
density=density.ravel(),
)
# Generate subspace-projected NUFFT operator
subspace_nufft = MRISubspace(nufft, subspace_basis=basis)
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
Generate K-Space#
We generate the k-space data using a non-projected operator. This can be simply obtained by using an identity matrix with of shape (ETL, ETL) (= number of contrasts) as a subspace basis.
multicontrast_nufft = [
get_operator("finufft")(
samples=samples[n],
shape=mri_data.shape[-2:],
density=density[n].ravel(),
)
for n in range(basis.shape[-1])
]
kspace = np.stack([multicontrast_nufft[n].op(mri_data[n]) for n in range(ETL)], axis=0)
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
/volatile/github-ci-mind-inria/gpu_mind_runner/_work/mri-nufft/venv/lib/python3.10/site-packages/mrinufft/_utils.py:94: UserWarning: Samples will be rescaled to [-pi, pi), assuming they were in [-0.5, 0.5)
warnings.warn(
Image reconstruction#
We now reconstruct both using the subspace expanded adjoint operator and the zero-filling adjoint operator followed by projection in image space
# Reconstruct with projection in image space
zerofilled_data_adj = np.stack(
[multicontrast_nufft[n].adj_op(kspace[n]) for n in range(ETL)], axis=0
)
zerofilled_display = (zerofilled_data_adj.T @ basis.T).T
zerofilled_display = np.concatenate(
[abs(coeff) / abs(coeff).max() for coeff in zerofilled_display], axis=-1
)
# Reconstruct with projection in k-space
subspace_data_adj = subspace_nufft.adj_op(kspace)
subspace_display = np.concatenate(
[abs(coeff) / abs(coeff).max() for coeff in subspace_data_adj], axis=-1
)
fig3, ax3 = plt.subplots(2, 1)
ax3[0].imshow(zerofilled_display, cmap="gray")
ax3[0].set_xticks([])
ax3[0].set_yticks([])
ax3[0].set(ylabel="kspace projection")
ax3[0].set_title("reconstructed subspace coefficients")
ax3[1].imshow(subspace_display, cmap="gray")
ax3[1].set_xticks([])
ax3[1].set_yticks([])
ax3[1].set(ylabel="zerofill + projection")
plt.show()
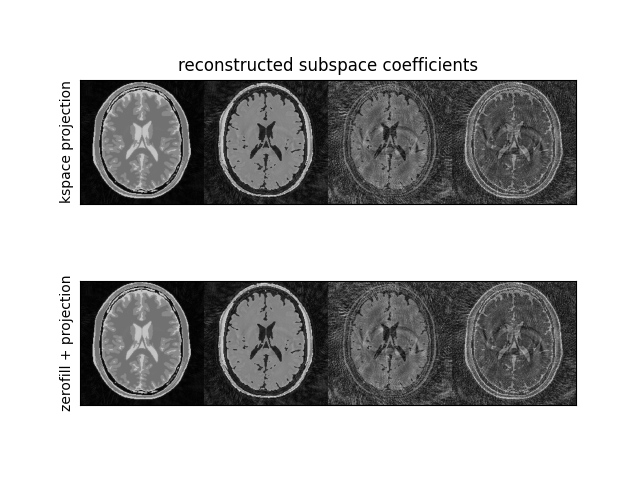
The projected k-space is equivalent to the regular reconstruction followed by projection.
Total running time of the script: (0 minutes 41.349 seconds)
