Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Off-resonance corrected NUFFT operator#
An example to show how to setup an off-resonance corrected NUFFT operator.
This example shows how to use the off-resonance corrected (ORC) NUFFT operator to reconstruct data in presence of B0 field inhomogeneities. Hereafter a 2D spiral trajectory is used for demonstration.
import matplotlib.pyplot as plt
import numpy as np
from mrinufft import display_2D_trajectory
plt.rcParams["image.cmap"] = "gray"
Data preparation#
Image loading#
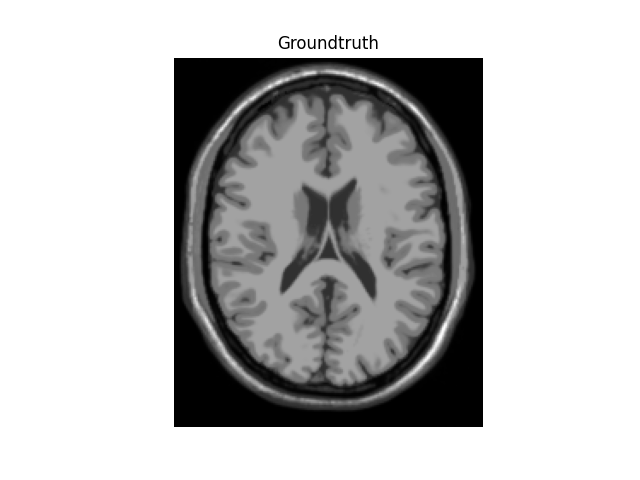
For realistic a 2D image we will use the BrainWeb dataset,
installable using pip install brainweb-dl.
plt.imshow(mri_data)
plt.axis("off")
plt.title("Groundtruth")
plt.show()
Mask generation#
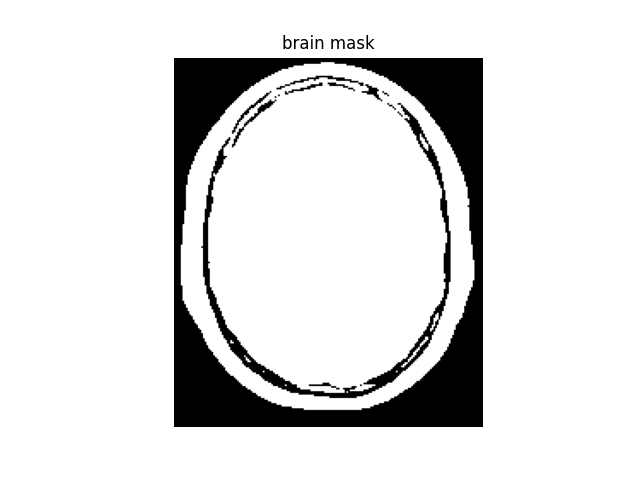
A binary mask is generated to exclude the background. We use a simple binary threshold for this example, but for real-world application it is advised to use more advanced methods and tools (e.g., FSL-BET).
brain_mask = mri_data > 0.1 * mri_data.max()
plt.imshow(brain_mask)
plt.axis("off")
plt.title("brain mask")
plt.show()
B0 field map generation#
A dummy B0 field map is generated for this example using the input shape.
from mrinufft.extras import make_b0map
b0map, _ = make_b0map(mri_data.shape, b0range=(-200, 200), mask=brain_mask)
plt.imshow(brain_mask * b0map, cmap="bwr", vmin=-200, vmax=200)
plt.axis("off")
plt.colorbar()
plt.title("B0 map [Hz]")
plt.show()
![B0 map [Hz]](../../_images/sphx_glr_example_offresonance_003.png)
Trajectory generation#
from mrinufft import initialize_2D_spiral
from mrinufft.density import voronoi
from mrinufft.trajectories.utils import DEFAULT_RASTER_TIME
samples = initialize_2D_spiral(Nc=48, Ns=600, nb_revolutions=10)
t_read = np.arange(samples.shape[1]) * DEFAULT_RASTER_TIME * 1e-3
t_read = np.repeat(t_read[None, ...], samples.shape[0], axis=0)
density = voronoi(samples)
Operator setup#
from mrinufft import get_operator
from mrinufft.operators.off_resonance import MRIFourierCorrected
# Generate standard NUFFT operator
nufft = get_operator("finufft")(
samples=2 * np.pi * samples, # normalize for finufft
shape=mri_data.shape,
density=density,
)
# Generate NUFFT off-resonance corrected operator
orc_nufft = MRIFourierCorrected(
nufft, b0_map=b0map, readout_time=t_read, mask=brain_mask
)
# Generate k-space
kspace_on = nufft.op(mri_data)
kspace_off = orc_nufft.op(mri_data)
# Reconstruct without B0 field inhomogeneity
mri_data_adj_ref = nufft.adj_op(kspace_on)
mri_data_adj_ref = np.squeeze(abs(mri_data_adj_ref))
# Reconstruct without B0 field correction
mri_data_adj = nufft.adj_op(kspace_off)
mri_data_adj = np.squeeze(abs(mri_data_adj))
# Reconstruct with B0 field correction
mri_data_adj_orc = orc_nufft.adj_op(kspace_off)
mri_data_adj_orc = np.squeeze(abs(mri_data_adj_orc))
/volatile/github-ci-mind-inria/gpu_runner2/_work/mri-nufft/venv/lib/python3.10/site-packages/finufft/_interfaces.py:336: UserWarning: Argument `data` does not satisfy the following requirement: C. Copying array (this may reduce performance)
warnings.warn(f"Argument `{name}` does not satisfy the following requirement: {prop}. Copying array (this may reduce performance)")
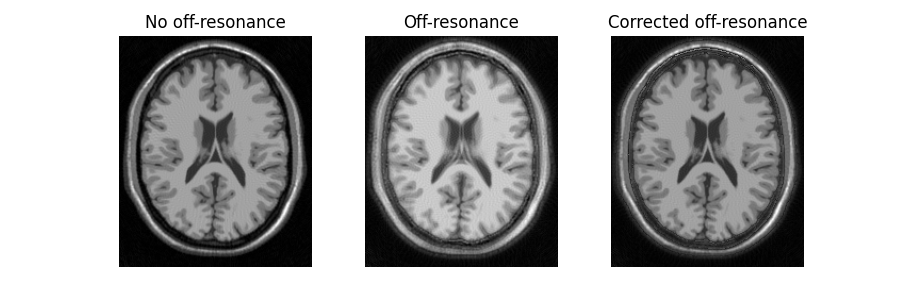
The blurring observed in the presence of B0 field inhomogeneities (middle) is significantly reduced using the off-resonance corrected NUFFT operator (right).
fig2, ax2 = plt.subplots(1, 3, figsize=(9, 3))
# No off-resonance
ax2[0].imshow(mri_data_adj_ref)
ax2[0].axis("off")
ax2[0].set_title("No off-resonance")
# No off-resonance correction
ax2[1].imshow(mri_data_adj)
ax2[1].axis("off")
ax2[1].set_title("Off-resonance")
# Off-resonance corrected
ax2[2].imshow(mri_data_adj_orc)
ax2[2].axis("off")
ax2[2].set_title("Corrected off-resonance")
plt.show()
Total running time of the script: (0 minutes 3.425 seconds)
